Research
Scientific interests:
- Molecular modeling for drug–design purposes
- Interactions of small drug–like molecules with macromolecular targets
- Dynamics of biological systems (RNA, proteins, modified oligomers)
- Allosteric mechanisms of enzyme activity
My projects:
- 2017 - 2022 (the last project) SONATA, Polish National Science Centre Computational investigation of dynamic and physico-chemical
properties of pteridine reductase 1 of human trypanosomatid
parasites, and its interactions with substrates and drugs. (grant no. 2016/21/D/NZ1/02806, role: principal investigator)
- 2017 - 2018 Grant from Center for Modelling and Simulation in the Biosciences BIOMS (Heidelberg University)
in collaboration with Heidelberg Institute for Theoretical Studies
- 2015 - 2016 EU project New Medicines for Trypanosomatidic Infections (postdoc at the Heidelberg Institute for Theoretical Studies - group of Prof. Rebecca Wade)
- 2011 - 2014 PRELUDIUM, Polish National Science Centre: Computational study of conformational and thermodynamic properties of human ribosomal A-site and its interactions with aminoglycoside antibiotics. (role: principal investigator)
- 2009 - 2012 TEAM, Foundation for Polish Science: Antisense peptide nucleic acids as inhibitors of bacterial translation (role: participant, PhD student)
The SONATA project (2017 - 2022)
Computational investigation of dynamic and physico-chemical properties of pteridine reductase 1 of human trypanosomatid parasites, and its interactions with substrates and drugs. (grant no. 2016/21/D/NZ1/02806)
Trypanosomatid parasites cause life–threatening human diseases such as leishmaniasis (Leishmania species), Chagas’ disease (Trypanosoma cruzi) and sleeping sickness (Trypanosoma brucei). Due to such problems as drug toxicity or parasite resistance, we need new more effective treatments for these infections. In particular, blocking the parasitic folate pathway, e.g., by repurposing the existing folate–pathway targeting drugs (antifolates), is one of the recently explored anti–trypanosomatid drug design strategies. However, for trypanosomatids, this can be only achieved by additionally inhibiting a trypanosomatid-specific protein, pteridine reductase 1 (PTR1) that causes resistance to antifolates, e.g., methotrexate.
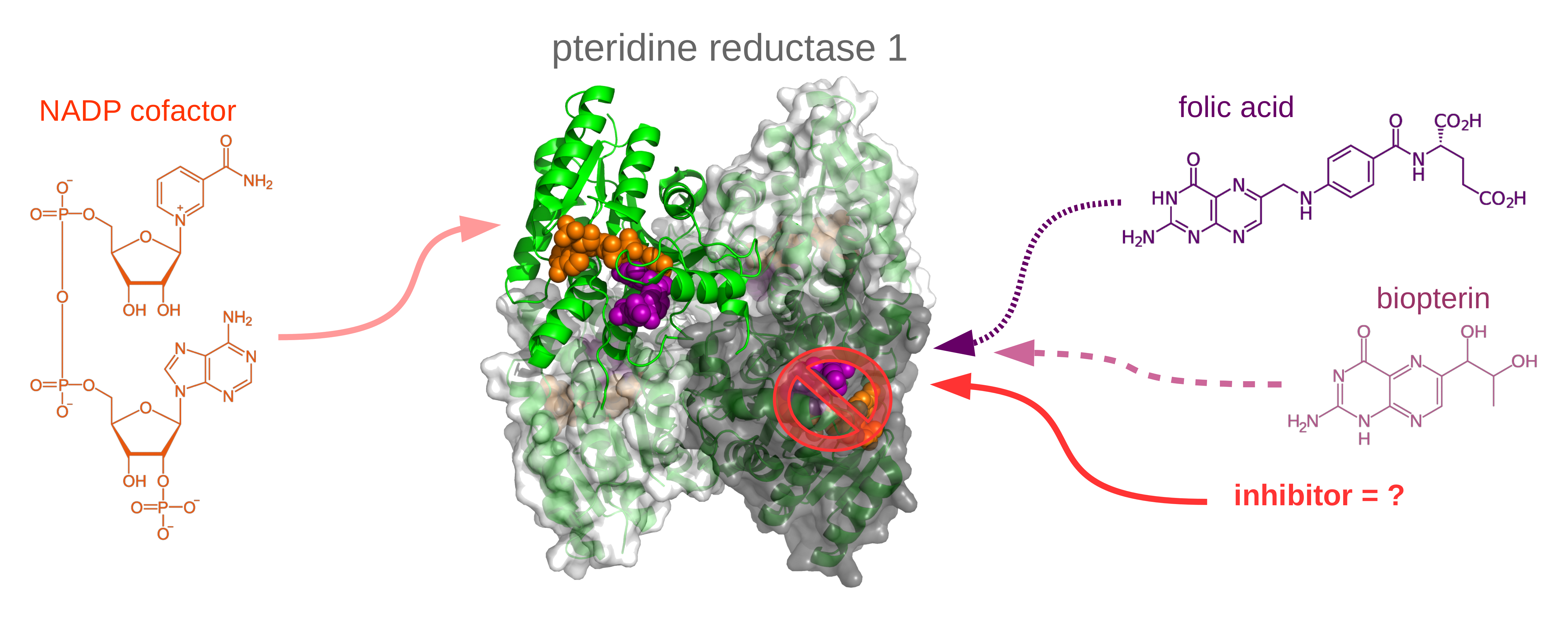
There is not much known about the molecular mechanisms that regulate the PTR1 activity. Biochemical experiments demonstrate interesting patterns of the PTR1 substrate specificity and also show that PTR1 is inhibited by semi-products of its catalytic reaction (substrate inhibition phenomenon); both phenomena are probably related to the physico-chemical and dynamic properties of PTR1. Furthermore, our preliminary studies suggested that understanding of the PTR1 dynamics may be also important for explaining differences in the inhibitor binding affinities. Since, in general, dynamics are known to play a crucial role in enzyme functioning, and the PTR1 dynamic properties have not been studied yet, we decided to investigate this aspect in the current project. We will employ computational methodology, in particular – molecular dynamics (MD) and related techniques. MD methods are used to study motions of the PTR1 protein, and how they are related to the PTR1 enzymatic activity and inhibitor binding. PTR1 performs its function as a homotetramer, which is a relatively big biomolecular system. Therefore, in order to explore large–scale movements of the PTR1 protein, we additionally employ enhanced sampling techniques.